The escalating crisis of antimicrobial resistance elevates the urgency for novel antibiotic agents to unprecedented levels. While the predominant sources of existing antibiotics are bacteria and fungi, the domain of Archaea presents a largely unexplored frontier for such discoveries. In a recent investigation, scientists affiliated with the University of Pennsylvania employed a sophisticated deep learning methodology to meticulously survey archaeal organisms. By analyzing the proteomes of 233 distinct archaeal species, they successfully pinpointed 12,623 molecular entities exhibiting potential antimicrobial efficacy.
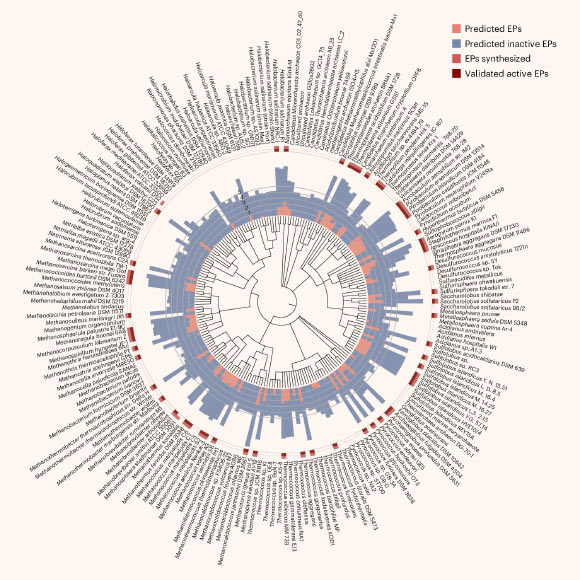
Torres et al. successfully synthesized 80 archaeasins, with a remarkable 93% demonstrating antimicrobial activity under laboratory conditions against problematic pathogens including Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, and various Enterococcus species. The visual representation is credited to Torres et al., with the corresponding digital object identifier being 10.1038/s41564-025-02061-0.
“Historically, the pursuit of new antibiotics has predominantly focused on sources such as fungi, bacteria, and animals,” remarked Dr. César de la Fuente, a distinguished researcher at the University of Pennsylvania.
“Previously, our application of artificial intelligence models facilitated the identification of promising antibiotic candidates from an array of unexpected origins, ranging from the genetic material of extinct organisms to the complex chemical compounds found in animal venoms.”
“Our current endeavor involves directing these advanced analytical tools toward a novel dataset: the protein profiles of a multitude of ancient microbial life forms.”
“Essentially, an entire distinct realm of biological existence remains ripe for comprehensive exploration.”
Archaea, occupying their own unique position on the evolutionary tree of life, are biologically distinct from both bacteria and eukaryotes, the latter encompassing plants, animals, and fungi.
Despite superficial similarities to bacteria when viewed microscopically, Archaea exhibit profound divergences in their genetic makeup, cellular membrane structures, and biochemical pathways.
These fundamental differences enable Archaea to flourish in some of the planet’s most extreme terrestrial and aquatic environments, including superheated subterranean ocean vents and scalding geothermal springs akin to those observed in Yellowstone National Park.
Given their inherent capacity to thrive in conditions prohibitive to most other life forms—withstanding immense hydrostatic pressure, exposure to toxic substances, and severe temperature fluctuations—the evolutionary trajectory of Archaea has led to the development of highly specialized biological mechanisms.
Consequently, they represent a promising yet largely unexploited reservoir of novel molecular agents, including compounds that may possess antibiotic-like properties but operate through mechanisms distinct from those of currently available antibiotics.
“Our interest in Archaea was initially piqued by their evolutionary imperative to develop sophisticated biochemical defense systems within their challenging habitats,” explained Dr. Marcelo Torres, also a member of the research team at the University of Pennsylvania.
“We hypothesized that their remarkable longevity and resilience over billions of years under such arduous conditions suggested the potential development of unique strategies for combating competing microorganisms, from which we might glean valuable insights.”
To identify potential antibiotic compounds embedded within the archaeal genome, the research team turned to cutting-edge artificial intelligence technologies.
They utilized an enhanced iteration of APEX, an artificial intelligence system originally engineered by the group for the discovery of antibiotic candidates within ancient biological samples, including the protein fragments of long-extinct fauna such as the woolly mammoth.
Having been trained on thousands of peptides—short chains of amino acids—with documented antimicrobial effects, APEX possesses the capability to accurately predict the likelihood of a given amino acid sequence exhibiting similar biological activity.
By reconfiguring APEX 1.1 with an expanded dataset comprising thousands of additional peptides and comprehensive information on bacteria that pose significant health threats to humans, the scientists refined the tool to forecast which archaeal peptides held the greatest potential for inhibiting bacterial proliferation.
The comprehensive analysis of 233 archaeal species led to the identification of over 12,000 potential antibiotic candidates.
These newly identified molecules were designated “archaeasins” by the study’s authors. Chemical characterization revealed that they possess distinct properties compared to established antimicrobial peptides (AMPs), notably in the differential distribution of their electrical charges.
Subsequently, a selection of 80 archaeasins was chosen for rigorous testing against live bacterial cultures.
“The conventional approach of searching for new antibiotics on a compound-by-compound basis is akin to an exhaustive search in an immense haystack,” stated Fangping Wan, a postdoctoral researcher at the University of Pennsylvania.
“Artificial intelligence significantly accelerates this process by efficiently pinpointing the most probable locations of such valuable discoveries.”
Antibiotics exert their effects through various mechanisms. Some disrupt the structural integrity of bacterial membranes, causing them to rupture, while others interfere with essential cellular processes, such as protein synthesis.
The researchers observed that, in contrast to the majority of known AMPs which primarily target the external defenses of bacteria, archaeasins appear to exert their influence from within the cell, disrupting the vital electrical signaling pathways crucial for cellular viability.
In empirical evaluations against a spectrum of antibiotic-resistant, pathogenic bacteria, an impressive 93% of the 80 archaeasins subjected to testing demonstrated measurable antimicrobial activity against at least one bacterial species.
The research consortium then proceeded to evaluate three selected archaeasins in preclinical animal models.
Following a single administration, the archaeasins effectively halted the propagation of a drug-resistant bacterium commonly implicated in hospital-acquired infections after a four-day observation period.
One of the three investigated compounds exhibited efficacy comparable to that of polymyxin B, an antibiotic frequently employed as a last resort against infections resistant to conventional treatments.
“This groundbreaking research underscores the substantial potential for discovering a multitude of antibiotics within the archaeal domain,” Dr. de la Fuente emphasized.
“With the escalating prevalence of bacteria developing resistance to existing antibiotics, it is of paramount importance to identify new therapeutic agents from unconventional sources to serve as crucial replacements.”
A detailed exposition of these findings has been published in the esteemed journal Nature Microbiology via the following link.
_____
Torres, M. D. T., et al. Deep learning reveals antibiotics in the archaeal proteome. Nat Microbiol, published online August 12, 2025; doi: 10.1038/s41564-025-02061-0