Newts, which are aquatic amphibians possessing tails, exhibit vast genomes that contain a significant abundance of repetitive DNA sequences. The precise influence of these repetitive elements on genomic architecture and their correlation with the remarkable regenerative capacities of newts remain subjects of scientific inquiry. In a recent investigation, scientists affiliated with the Karolinska Institute, alongside other institutions, have successfully generated a chromosome-level genomic sequence for the Iberian ribbed newt (Pleurodeles waltl).
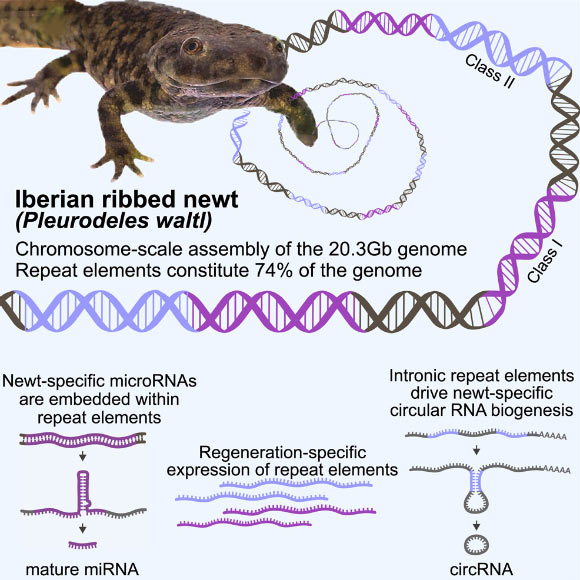
Brown et al. have presented a chromosome-scale assembly of the 20.3 Gb genome belonging to the Iberian ribbed newt (Pleurodeles waltl), characterized by unparalleled contiguity and completeness among genomes of this magnitude. Provided courtesy of Brown et al., with the DOI: 10.1016/j.xgen.2025.100761.
This species, also recognized as the gallipato or Spanish ribbed newt, is indigenous to Spain, Portugal, and Morocco.
The defining characteristics of this newt species include a broad, flattened cranial structure and prominent ribs that have the capacity to protrude through its flanks.
Mature males can achieve lengths of up to 31 cm (12.2 inches), while females typically reach a size of 29 cm (11.4 inches). Notably, individuals originating from North Africa tend to be smaller in stature compared to their European counterparts.
“The Iberian ribbed newt exhibits an extraordinary capacity for regeneration, possessing the ability to regrow lost appendages and repair damaged tissues within complex organs such as the brain, heart, and eyes,” stated Professor András Simon of the Karolinska Institute, along with his research collaborators.
“The utility of this valuable model organism would be substantially augmented by the availability of a high-fidelity genome assembly and annotation.”
“However, this endeavor has presented considerable challenges, predominantly due to the substantial size of its genome, approximately 20 Gb, and the significant prevalence of repetitive DNA sequences.”
The researchers’ analysis revealed that repetitive elements constitute a substantial 74% of the Iberian ribbed newt’s genomic makeup.
“While this presented a significant technical hurdle, we achieved success through meticulous mapping efforts that surpass previous efforts in comprehensiveness for any species with a comparable genome size,” Professor Simon elaborated.
“We precisely established the locations of both protein-coding and non-coding genetic sequences on each individual chromosome,” explained Ketan Mishra, a Ph.D. student at the Karolinska Institute.
“Furthermore, we identified specific protein-coding genes that are either absent from the newt’s genome or present in amplified copy numbers relative to other species.”
“These findings represent a critical foundational resource for researchers across diverse scientific disciplines, including evolutionary genomics, developmental and regenerative biology, and cancer research.”
“The subsequent phase of our research will involve undertaking functional studies, wherein we will systematically perturb molecular processes to ascertain their impact on regenerative capabilities.”
“Concurrently, we are planning to conduct comparative analyses with other species to deepen our understanding of these intricate regenerative mechanisms.”
The results of this investigation have been published in the esteemed journal Cell Genomics.
_____
Thomas Brown et al. Chromosome-scale genome assembly reveals how repeat elements shape non-coding RNA landscapes active during newt limb regeneration. Cell Genomics, published online January 27, 2025; doi: 10.1016/j.xgen.2025.100761