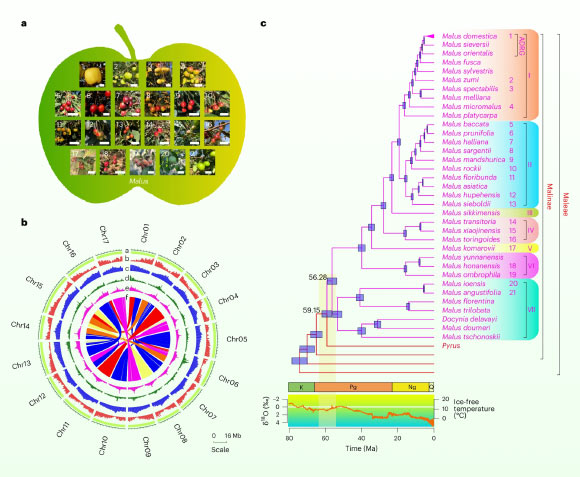
The genus Malus encompasses over 35 distinct species, with a natural distribution spanning the temperate regions of the Northern Hemisphere, from Eastern Asia and Europe to North America. This botanical group includes the cultivated apple, scientifically known as Malus domestica, alongside its wild progenitors. Recent scientific investigations have shed light on the intricate evolutionary connections between various Malus species and the developmental trajectory of their genetic blueprints over the last approximately 60 million years.
The
Malus
evolutionary landscape, as depicted through phylogenomics. Image courtesy of Li
et al
., doi: 10.1038/s41588-025-02166-6.
“While the genus Malus is comprised of approximately 35 species, a comprehensive understanding of the evolutionary pathways of their genomes has been notably absent, despite the significant agricultural importance of apples,” remarked Penn State Professor Hong Ma.
“Our research facilitated an in-depth examination of Malus genomes, enabling the construction of a phylogenetic framework for apples. This allowed us to meticulously document key evolutionary events, such as whole-genome duplications and interspecies hybridization. Furthermore, we identified specific genomic regions correlated with agriculturally relevant traits, including resistance to apple scab pathology.”
Professor Ma and his research associates undertook the task of newly sequencing and assembling the complete genomes of 30 members of the Malus genus, which included the well-known golden delicious apple cultivar.
Among the 30 studied species, 20 exhibit diploid chromosomal arrangements, meaning they possess two sets of each chromosome, analogous to human genetics. The remaining 10 species are polyploid, characterized by three or four sets of chromosomes. This polyploidy is likely the result of relatively recent hybridization events involving diploid individuals and other related species within the Malus genus.
By undertaking a comparative analysis of nearly 1,000 genes from each species, the research team successfully constructed an evolutionary family tree for the genus. Subsequent biogeographical analysis permitted the tracing of the genus’s origins back to approximately 56 million years ago in Asia.
“The evolutionary narrative of this genus is remarkably intricate, marked by frequent instances of interspecies hybridization and a shared whole-genome duplication event that introduces complexities in comparative analyses,” Professor Ma elaborated.
“The availability of high-fidelity genomes for a substantial proportion of the genus’s species, coupled with a clear understanding of their interrelationships, empowered us to delve more profoundly into the mechanisms driving the genus’s evolutionary progression.”
To further scrutinize the historical development and evolutionary patterns of the Malus genomes, the scientists employed an analytical methodology known as pan-genomics.
This approach involved a thorough comparison of both conserved genetic elements and other sequence types, including transposons—often referred to as “jumping genes” due to their mobile nature within the genome—across the 30 analyzed genomes. It also accounted for genes exclusive to specific subsets of these genomes.
Pan-genomic analyses integrate genomic data from a group of closely related organisms to elucidate evolutionary conservation and divergence patterns. The efficacy of these analyses was significantly enhanced by the utilization of a pan-genome graph tool.
“Leveraging the pan-genome of 30 species proved exceptionally effective in identifying structural variations, as well as gene amplifications and rearrangements, among the species that might otherwise be overlooked in studies involving the comparison of only a few genomes,” Professor Ma stated.
“In this specific investigation, one identified structural variant enabled us to precisely locate the genomic segment responsible for conferring resistance to apple scab, a prevalent fungal affliction impacting apples globally.”
The research team also developed a specialized pan-genome analysis tool designed to detect evidence of selective sweeps, a process wherein a beneficial genetic trait rapidly proliferates within a population.
Employing this advanced method, they were able to pinpoint a genomic region associated with enhanced cold tolerance and disease resistance in wild Malus species. This same region was also found to potentially correlate with less palatable fruit characteristics.
“It is plausible that the drive to cultivate apples with superior gustatory qualities may have inadvertently compromised the hardiness of domesticated apple varieties,” Professor Ma hypothesized.
“An in-depth comprehension of the structural variations within Malus genomes, the intricate relationships among its species, and their hybridization history, as elucidated by pan-genome analysis, could provide invaluable guidance for future breeding initiatives. This would aim to simultaneously preserve desirable traits for enhanced flavor and robust disease resistance in apples.”
The comprehensive results of this groundbreaking research have been formally published in the esteemed scientific journal Nature Genetics.
_____
W. Li et al. Pan-genome analysis reveals the evolution and diversity of Malus. Nat Genet, published online April 16, 2025; doi: 10.1038/s41588-025-02166-6